The Lens Platform provides a suite of whole slide image analysis tools, including detection and segmentation of pathology structures,
such as nuclei, cells, clumped histology objects, vessels, and registration of serial slides from various modalities.
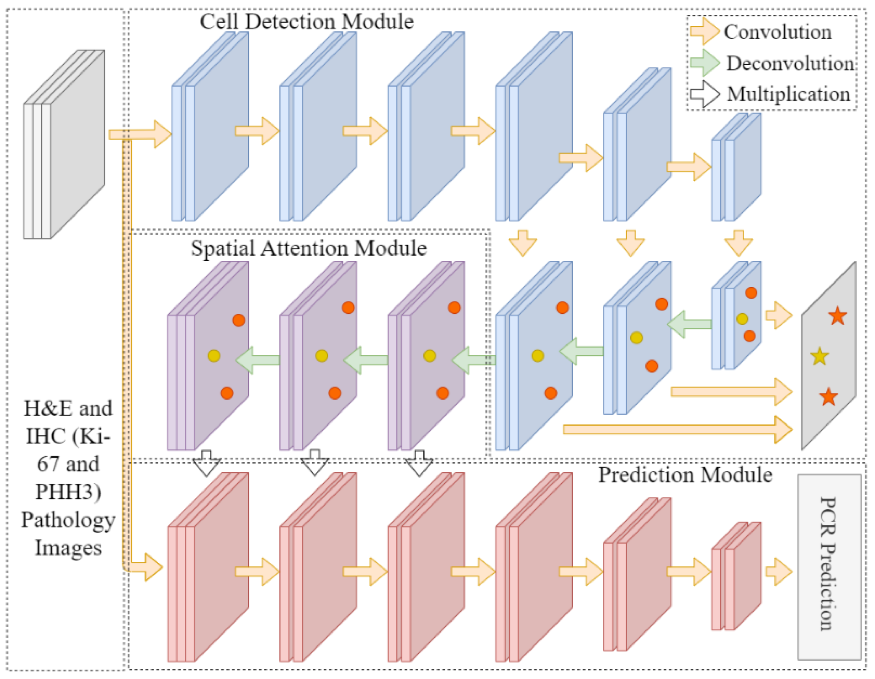
In triple negative breast cancer (TNBC) treatment, early prediction of pathological complete response (PCR) from chemotherapy before surgical operations is crucial for optimal treatment planning. We propose a novel deep learning-based system to predict PCR to neoadjuvant chemotherapy for TNBC patients with multi-stained histopathology images of serial tissue sections. By first performing tumor cell detection and recognition in a cell detection module, we produce a set of feature maps that capture cell type, shape, and location information. Next, a newly designed spatial attention module integrates such feature maps with original pathology images in multiple stains for enhanced PCR prediction in a dedicated prediction module. We compare it with baseline models that either use a single-stained slide or have no spatial attention module in place. Additionally, the heatmaps generated from the spatial attention module can help pathologists in targeting tissue regions important for disease assessment. Our system presents high efficiency and effectiveness and improves interpretability, making it highly promising for immediate clinical and translational impact.
Source code:
https://github.com/jkonglab/PCR_Prediction_Serial_WSIs_cells
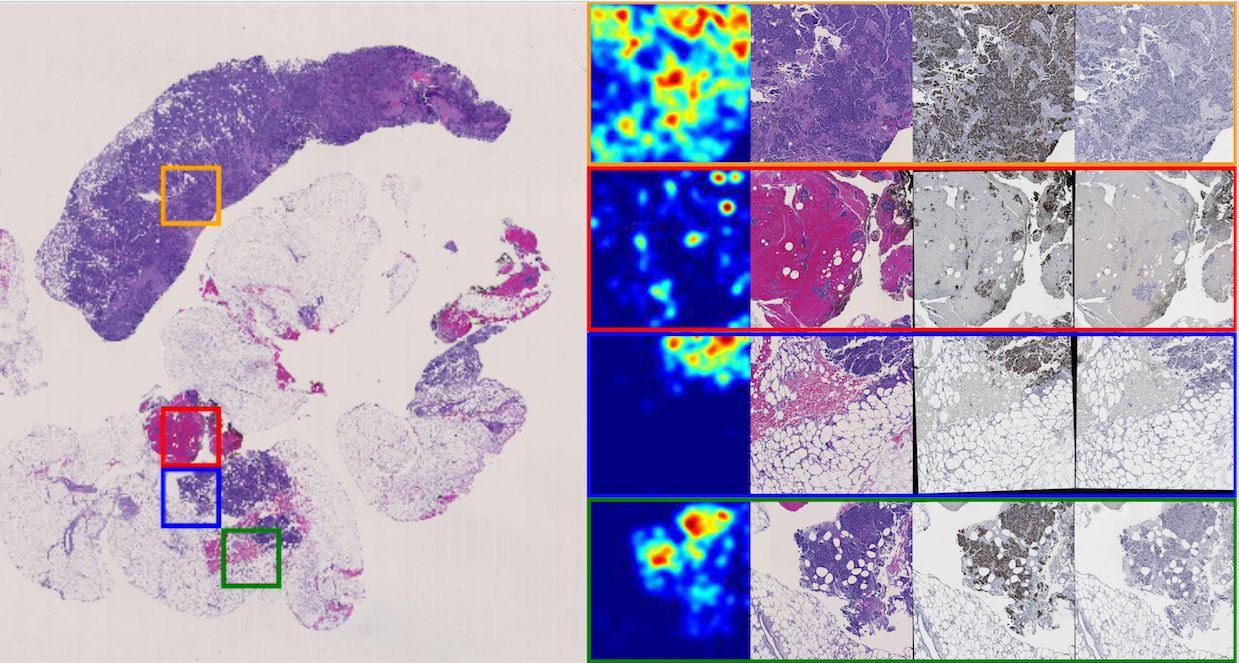
The pathological complete response to neoadjuvant chemotherapy is one good assessment measure in breast cancer treatment that is proven to be highly associated with patients' overall survival. While the accurate early prediction of PCR before neoadjuvant chemotherapy treatment is in high demand, the existing PCR prediction systems are too limited in accuracy to be deployed in real clinical settings. In this work, we proposed one convolutional neural network based system for predicting PCR from three different modalities of pathology images (hematoxylin and eosin (H&E), KI-67, and PHH3 stained) captured from adjacent tissue slices. One spatial attention mechanism is deployed to the system to guide it to pay attention to specific areas where pathologists consider important and informative. With integrated usage of different image modalities, the system can avoid the limitation of spatial homogeneity from original CNN. Our proposed model achieved a high patient-level accuracy, outperforming baselines and other state-of-the-art systems. Its inspiring performance suggests its great potential in real clinical usage.
Source code:
https://github.com/jkonglab/PCR_Prediction_Serial_WSIs_biomarkers
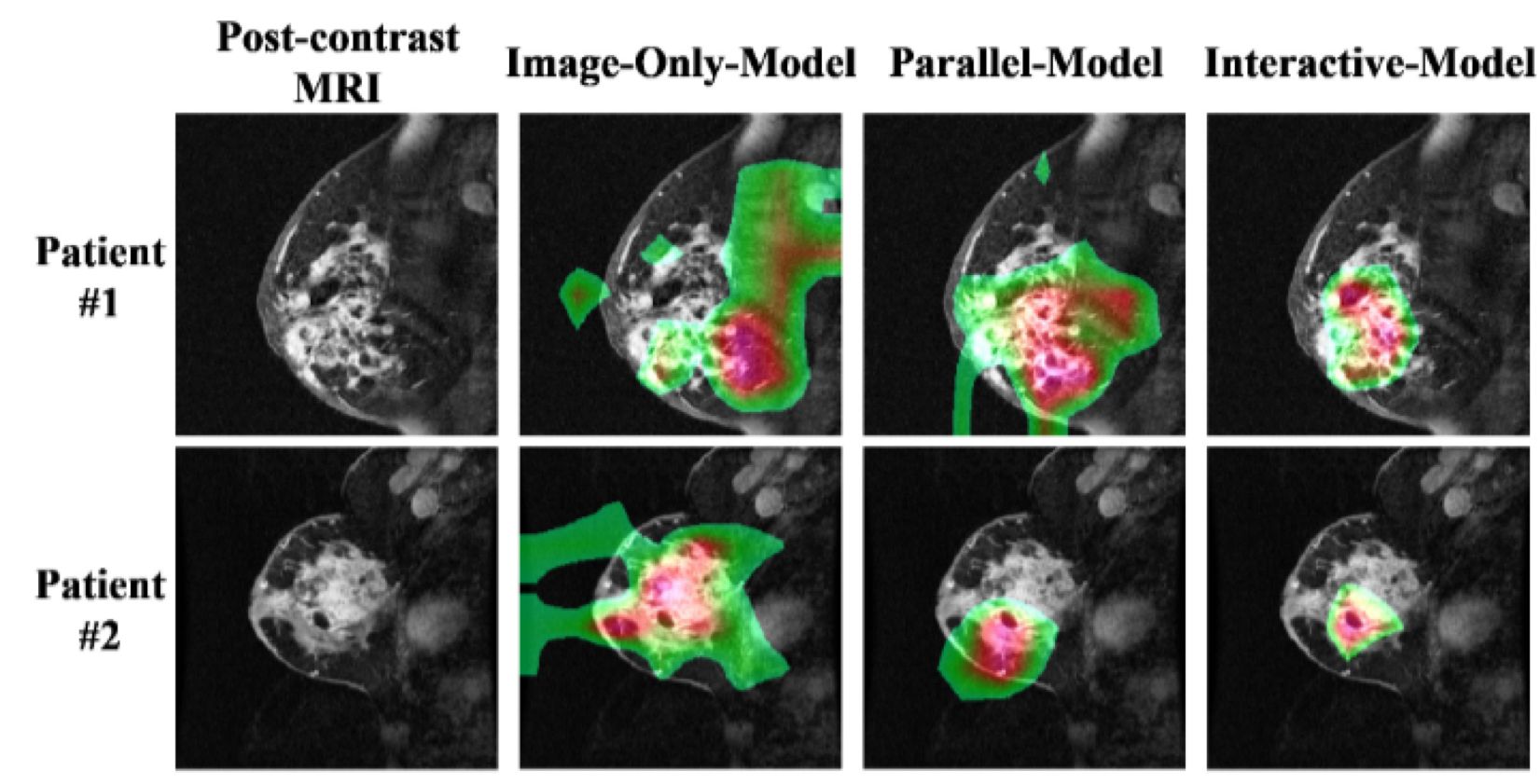
Neoadjuvant chemotherapy is widely used to reduce tumor size to make surgical excision manageable and to minimize distant metastasis. Assessing and accurately predicting pathological complete response is important in treatment planning for breast cancer patients. We propose a novel approach integrating 3D MRI imaging data, molecular data and demographic data using convolutional neural network to predict the likelihood of pathological complete response to neoadjuvant chemotherapy in breast cancer. We take post-contrast T1-weighted 3D MRI images without the need of tumor segmentation, and incorporate molecular subtypes and demographic data. In our predictive model, MRI data and non-imaging data are convolved to inform each other through interactions, instead of a concatenation of multiple data type channels. This is achieved by channel-wise multiplication of the intermediate results of imaging and non-imaging data. Our model significantly outperforms models using imaging data only or traditional concatenation models. Our approach has the potential to aid physicians to identify patients who are likely to respond to neoadjuvant chemotherapy at diagnosis or early treatment, thus facilitate treatment planning, treatment execution, or mid-treatment adjustment.
Source code:
https://github.com/jkonglab/PCR_Prediction_MRI_Molecular_Demographics