
Significant advancements in high-throughput tissue scanning technologies have enabled the imaging and analysis of tissues at cellular and subcellular levels of detail. A large number of studies using data derived from such high-resolution tissue images have produced characterizations of tumor microenvironments and tissue structure profiles that mirror molecular tumor signatures and disease progression patterns. In the oncology setting, histopathologic microscopy imaging data has become a prominent data modality that is critical for extreme-scale quantitative disease progression mechanism analyses, as well as disease diagnosis and prognosis. To capture such high levels of tissue structure detail, modern high-throughput scanners produce gigapixel-sized images of whole-slide tissue sections at a high resolution, typically at 40x objective magnification. The formidable size of the data captured in these high-resolution images has presented challenges in data analysis feasibility and performance. Traditional manual analyses of pathologic hallmarks have often become unfeasible due to the gigapixel image size and the high degree of inter- and intra-reader variability in this qualitative process. Therefore, automated computerized image analysis algorithms have emerged in the field of digital pathology as a means to more accurately assess pathologic features captured in high-resolution tissue images that may be relevant to diagnosis or prognosis in clinical settings.
The challenges in working with such large-scale image files make web and distributed cloud-based applications a suitable alternative for histopathology image storage, management, and analysis. First, web applications have high availability and are accessible from any modern web-enabled device without any prior installation or configuration. Second, web-based frameworks facilitate the storage and portability of large amounts of data, as uploaded files are stored in a central cloud server, reducing the storage burden of individual users and the need for sustained local storage of large datasets. Large amounts of data can be made accessible to others without actually copying or transferring datasets. Likewise, the obstacle of sharing large files with collaborators is no longer an issue, as data can be shared within seconds via access-granting links. Last and most importantly, with a web application, users do not need powerful local computational resources. A remote web application architecture allowing users to access a shared cluster of computational facilities can help address the computational challenges in whole-slide histopathology image analysis.
Another factor limiting full utilization of microscopy data is that almost all current tissue-based studies and commercial microscopy image analysis tools are constrained by the two-dimensional (2D) image space, causing significant loss of morphology and spatial information from the intrinsic tissue space. Three-dimensional (3D) microscopes were recently proposed to represent the information-lossless 3D tissue space by 3D microscopy image volumes. This novel concept has significant potential to enhance cancer research, as a large set of cancer biology research has demonstrated the importance of 3D histopathology features in the diagnosis of clinical outcomes. These 3D histopathology features include 3D structure volume scale, morphology features, spatial organizations, and alignment directionality.
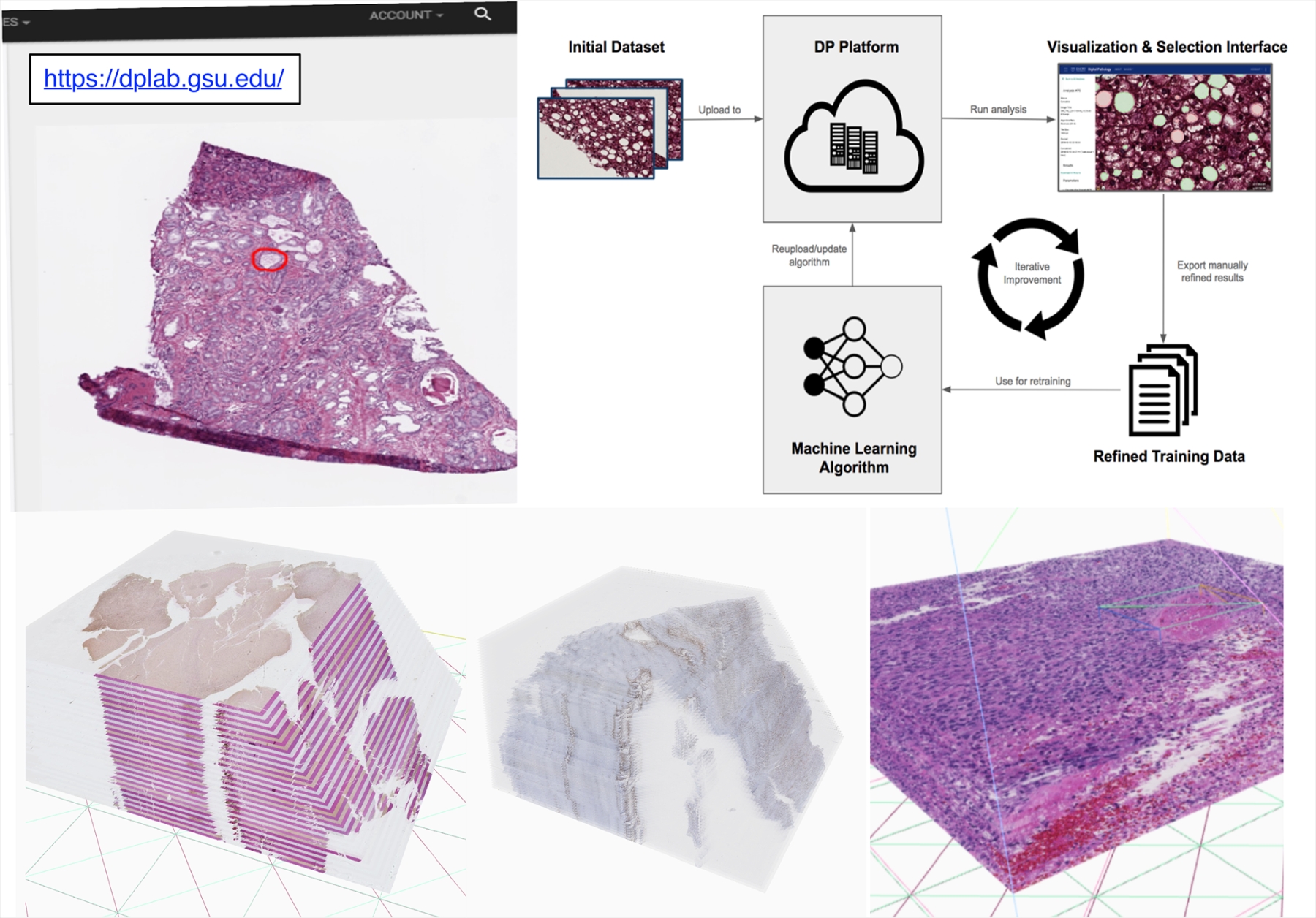
To address limitations above, we have been actively developing Digital Pathology Laboratory (dpLAB) as an online workbench to facilitate the end-to-end analysis pipeline of pathology image data in a variety of formats and modalities, including 2D, 3D, and spectroscopy microscopy images. Using dpLAB, users can upload and store their image data in a cloud-based server, remotely run computationally-expensive analysis algorithms using a distributed server cluster and export and share their results for further use. Users can either upload their algorithms or use dpLAB analysis algorithm library as a code-free, and "wrapper" web interface, thereby encouraging usage, distribution, and validation of new algorithms in both research and clinical contexts. The platform addresses critical needs in the field of digital pathology by providing a publicly-accessible data analysis and management web service to the research and clinical community at large. We intend to continue the development of dpLAB by providing more analysis integration features across imaging, clinical, and genomic data, expanding the library of available open-source algorithms, and improving support for more programming languages and image formats.
We have prototyped a publicly available web computing platform. This online platform creates a lab-in-a-browser solution that can be used by teams of researchers and clinicians to upload, manage, annotate, share, and visualize image data as well as perform experiments and run image analyses directly within a web browser. In this project, we will incorporate all Lens Platform software tools into the platform, and extend the storage and computational power to support pilot studies from the community. Users can submit a request to evaluate the Lens Platform with new pilot studies; once approved, users will be able to upload their own data based on instructions and constraints.
Demo:3D WSI Visualization: https://dplab.gsu.edu/demo
Full Web Application: https://dplab.gsu.edu
Source code: