The Lens Platform provides a suite of whole slide image analysis methods, including detection and segmentation of pathology structures,
such as nuclei, cells, clumped histology objects, vessels, and registration of serial slides from various modalities.
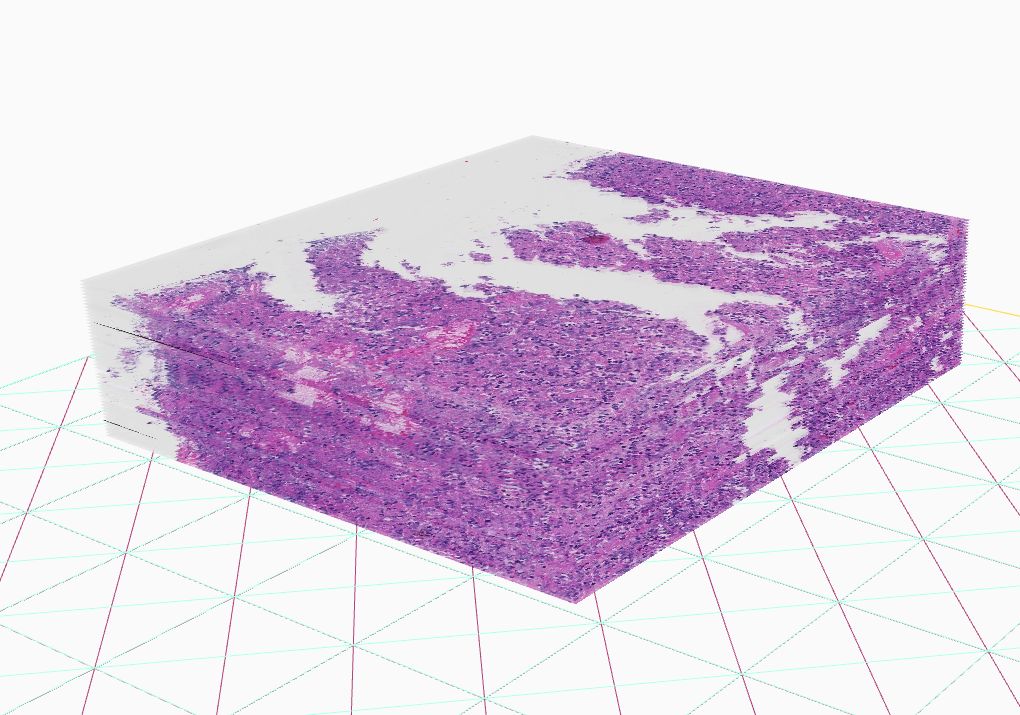
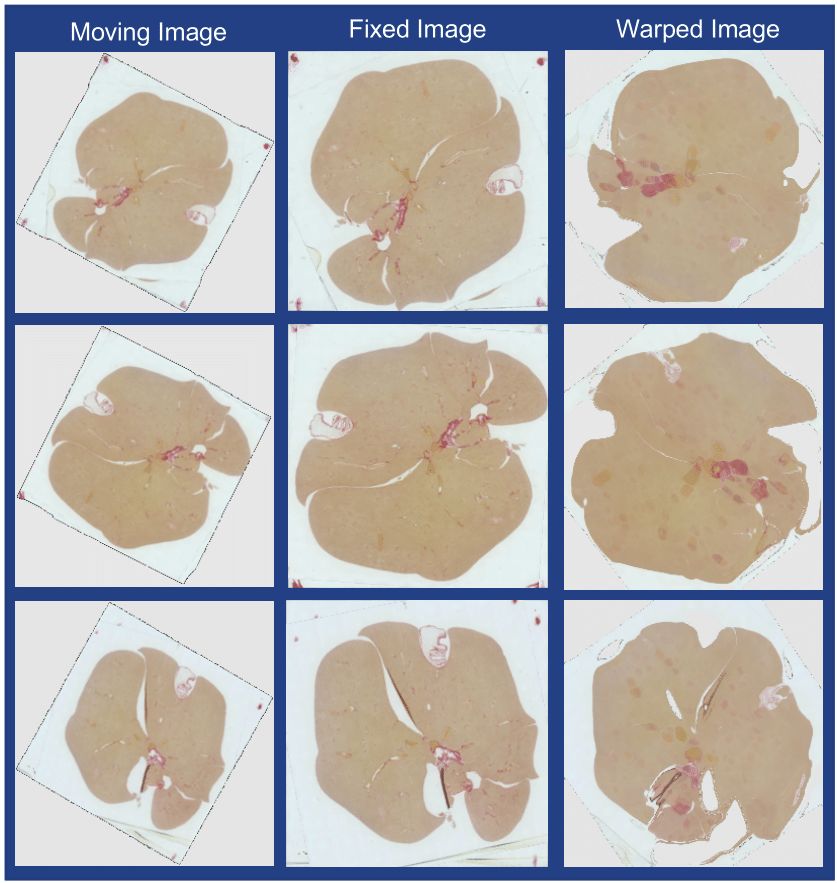
The introduction of high-throughput scanning technology has allowed for routine digital 3D reconstruction of serial whole slide histology images. However, registration of serial whole slide images is a challenge due to the overwhelmingly large image size. A single image may contain several gigabytes of data. The resulting 3D volumes can easily exceed modern computer memory limits, thus precluding the direct use of existing reconstruction methods. Based on current methods, accessing subvolumes of tissues requires the prior registration of the entire full-resolution whole slide image volume, leading to a high computational cost. To solve this problem, we develop a hierarchical image registration method that computes image deformation on-the-fly. In this way, we can minimize the computational burden and focus analysis on the tissue subvolumes of interest to domain experts. As the nonrigid registration at the low resolution is applied to the rigidly registered image volume and only alignment transformations for all adjacent image pairs are available, we have developed a novel recursive mapping method that maps each pixel in the reference coordinate system to the corresponding location in a target image with use of the mapped nonrigid and rigid propagated transformations from low to high image resolution. In addition, we are also developing a multi-resolution, block-wise registration approach that works in a coarse-to-fine manner for multi-stain image registration.
Source code: https://github.com/jkonglab/DigitalPathology
Demo: https://math.gsu.edu/jkong/research_registration.html

We have proposed a novel translation based fully unsupervised synthetic mono-modal image registration approach for serial whole-slide microscopy images in different stains. The registration approach is unsupervised since no prior deformation field information is needed for training the deep learning models. We have compared our method performance with multiple standard deep learning based models and demonstrated our method superiority in both quantitative and qualitative manner.
Source code: https://github.com/jkonglab/MultiStain_WSI_Registration_CycleGAN
3D digital pathology has been emerging for next-generation tissue based cancer re- search. To enable such histopathology image volume analysis, serial histopathology slides need to be well aligned. We propose a histopathology image registration fine tuning method with integrated landmark evaluations by texture and spatial proximity measures. Representative anatomical structures and image corner features are first detected as landmark candidates. Next, we identify strong and modify weak matched landmarks by leveraging image texture features and landmark spatial proximity measures. Both qualitative and quantitative results of extensive experiments demonstrate that our proposed method is robust and can further enhance registration accuracy of our previously registered image set. The promising experimental results suggest that our method can be used as a fine tuning module to further boost registration accuracy, a premise of histology spatial and morphology analysis in an information-lossless 3D tissue space for cancer research.
Source code: https://github.com/jkonglab/LandMark_Registration
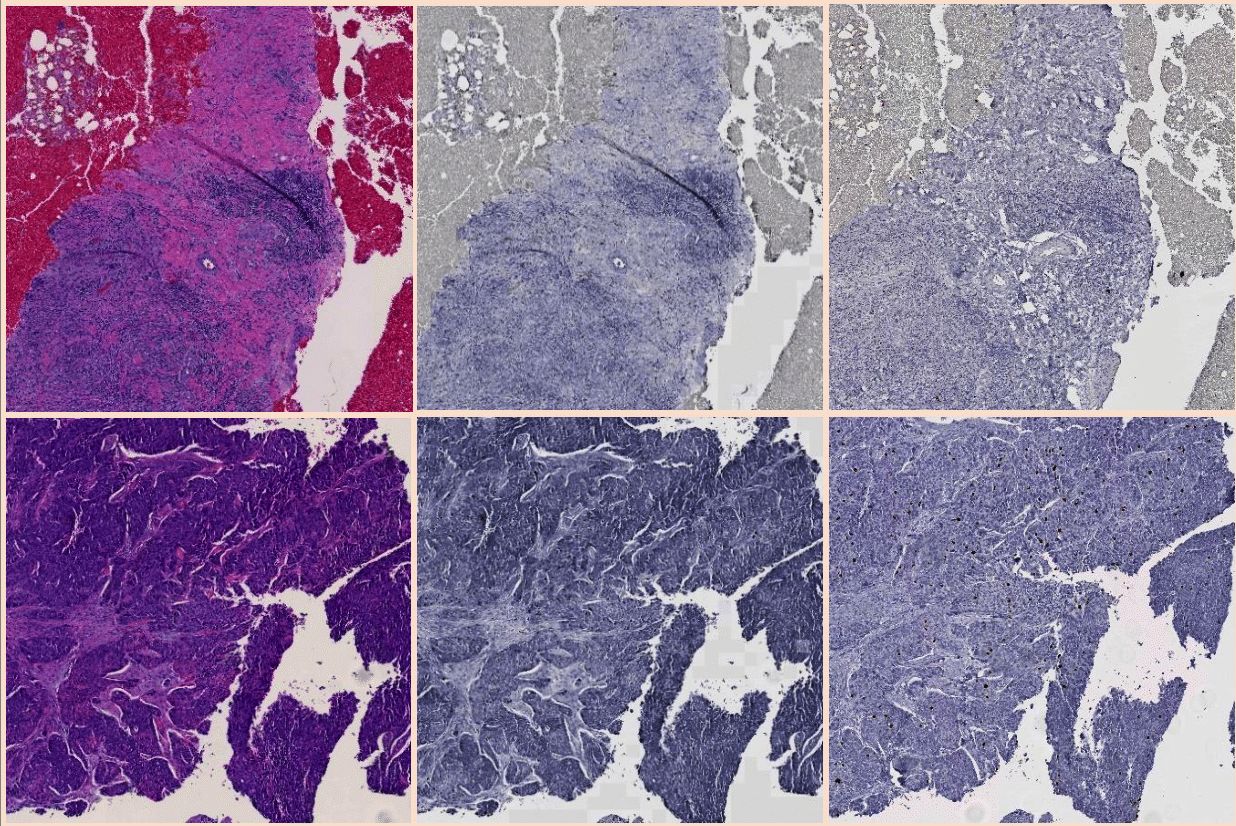
Registration of whole-slide Immunohistochemistry (IHC) images is an essential prerequisite for serial slide integra- tion analysis. However, the overwhelming size and artifacts in such images make the registration process challenging. We have developed HistoRegNet, an end-to-end unsuper- vised patch-based deep learning registration model, to spatially align IHC histopathology images. For efficacy demonstration, HistoRegNet is trained with image patches from 50 IHC whole-slide images that highlight mouse liver fibrosis tissues by the Sirius red stain. For model training, we use Normalized Cross-Correlation (NCC) as the similarity metric. The performance of HistoRegNet is com- pared with multiple state-of-the-art conventional as well as deep leaning methods by both image similarity measures (e.g. NCC, and normalized mutual information) and region over- lap measures such as Dice coefficient. Experimental results demonstrate the superior performance of our proposed model to other methods, suggesting its promising potential for IHC histopathology image registration.
Source code: coming soon...
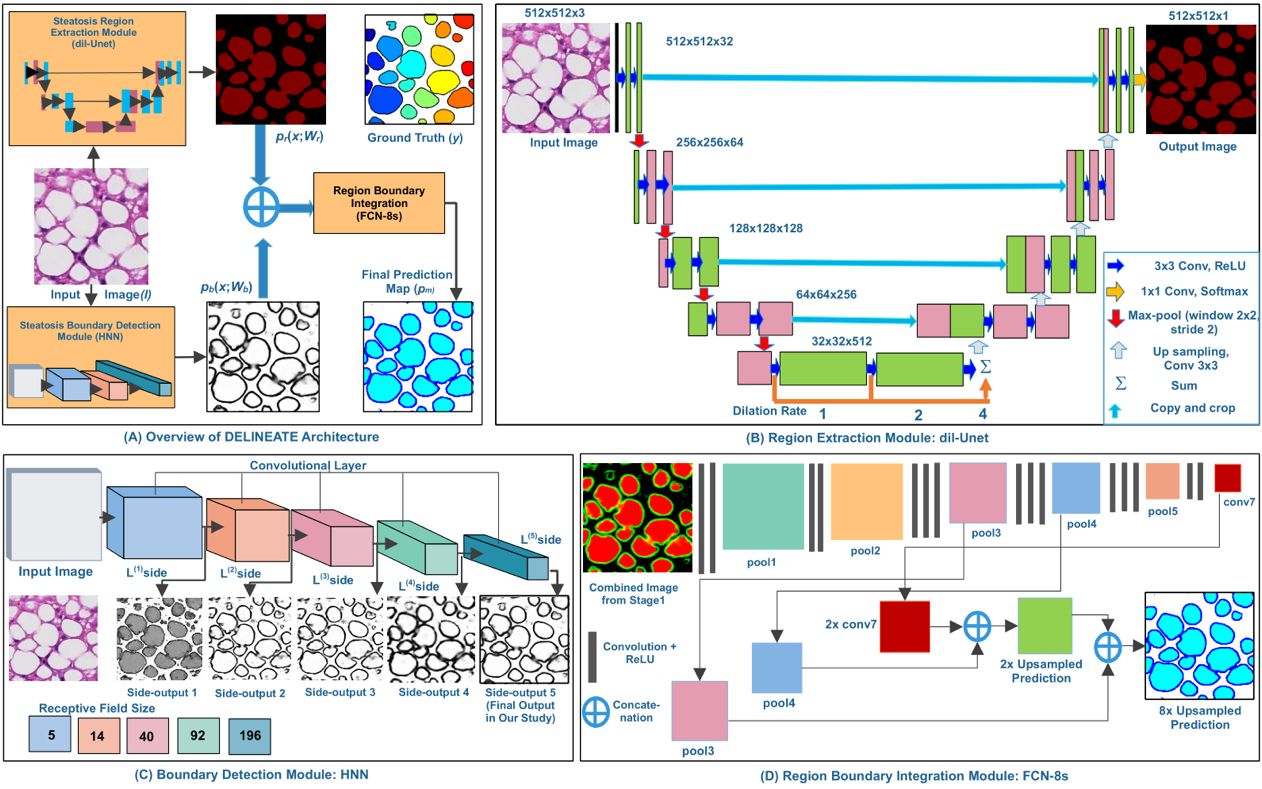
Hepatic steatosis droplet quantification with histology biopsies has high clinical significance for risk stratification and management of patients with fatty liver diseases and in the decision to use donor livers for transplantation. However, pathology reviewing processes, when conducted manually, are subject to a high inter- and intra-reader variability, due to the overwhelmingly large number and significantly varying appearance of steatosis instances. Meanwhile, this process is challenging as there is a large number of overlapped steatosis droplets with either missing or weak boundaries. We propose a deep learning based region-boundary integrated network for precise steatosis quantification with whole slide liver histopathology images. The proposed model consists of two sequential steps: a region extraction and boundary prediction module for foreground regions and steatosis boundary prediction, followed by an aggregated prediction map generation. Missing steatosis boundaries are next recovered from the predicted map and assembled from adjacent image patches to generate results for the whole slide histopathology image. The resulting steatosis measures at both the pixel level and steatosis object level present strong correlation with pathologist annotations, radiology readouts and clinical data. In addition, the count of isolated steatosis objects is shown as a promising alternative measure to the traditional metrics at the pixel level. These results suggest a high potential of AI assisted technology to enhance liver disease decision support using whole slide images.
Source code:
https://github.com/jkonglab/DELINEATE
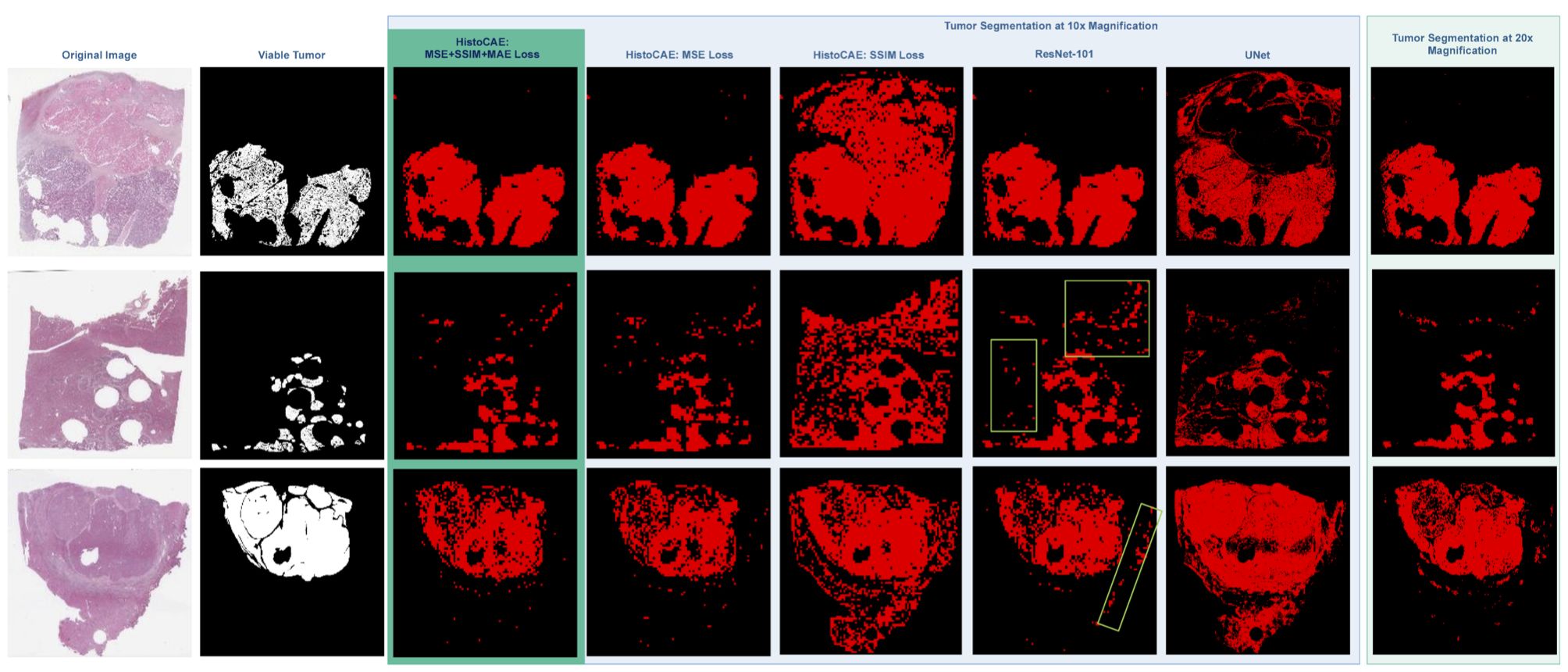
In this study, we present a multi-resolution deep learning model HistoCAE for viable tumor segmentation in whole-slide liver histopathology images. We propose convolutional autoencoder (CAE) based framework with a customized reconstruction loss function for image reconstruction, followed by a classification module to classify each image patch as tumor vs non-tumor. The resulting patch-based prediction results are spatially combined to generate the final segmentation result for each WSI. Our proposed model presents superior performance to other benchmark models with extensive experiments, suggesting its efficacy for viable tumor area segmentation with liver whole-slide images.
Source code:
https://github.com/jkonglab/Liver_Cancer_Segmentation
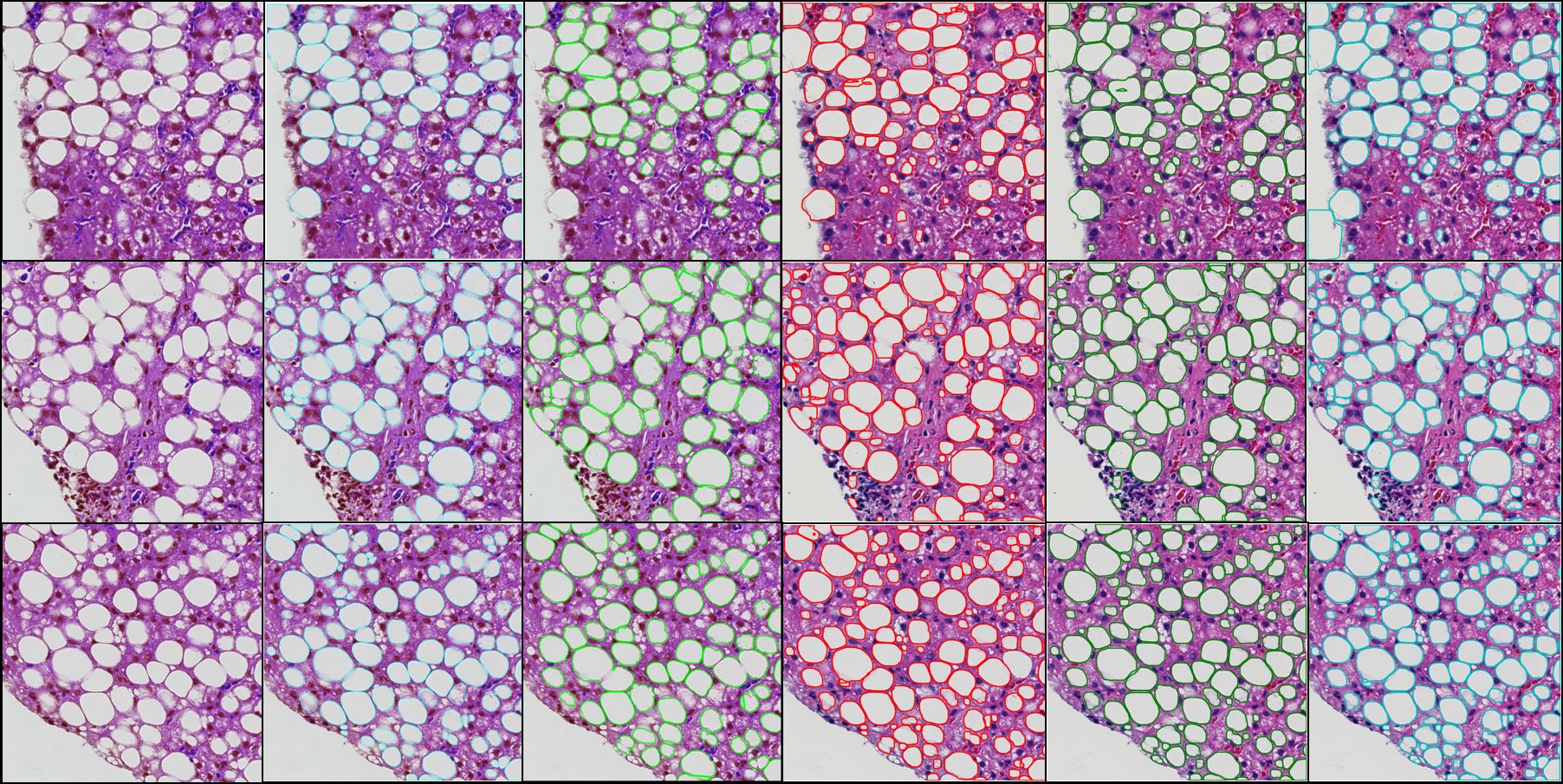
Liver steatosis is known as the abnormal accumulation of lipids within cells. An accurate quantification of steatosis area within the liver histopathological microscopy images plays an important role in liver disease diagnosis and trans- plantation assessment. Such a quantification analysis often requires a precise steatosis segmentation that is challenging due to abundant presence of highly overlapped steatosis droplets. In this paper, a deep learning model Mask-RCNN is used to segment the steatosis droplets in clumps. Extended from Faster R-CNN, Mask-RCNN can predict object masks in addition to bounding box detection. With transfer learning, the resulting model is promising to support liver disease diagnosis and allograft rejection prediction in future clinical practice.
Source code:
https://github.com/jkonglab/Steatosis_Segmentation
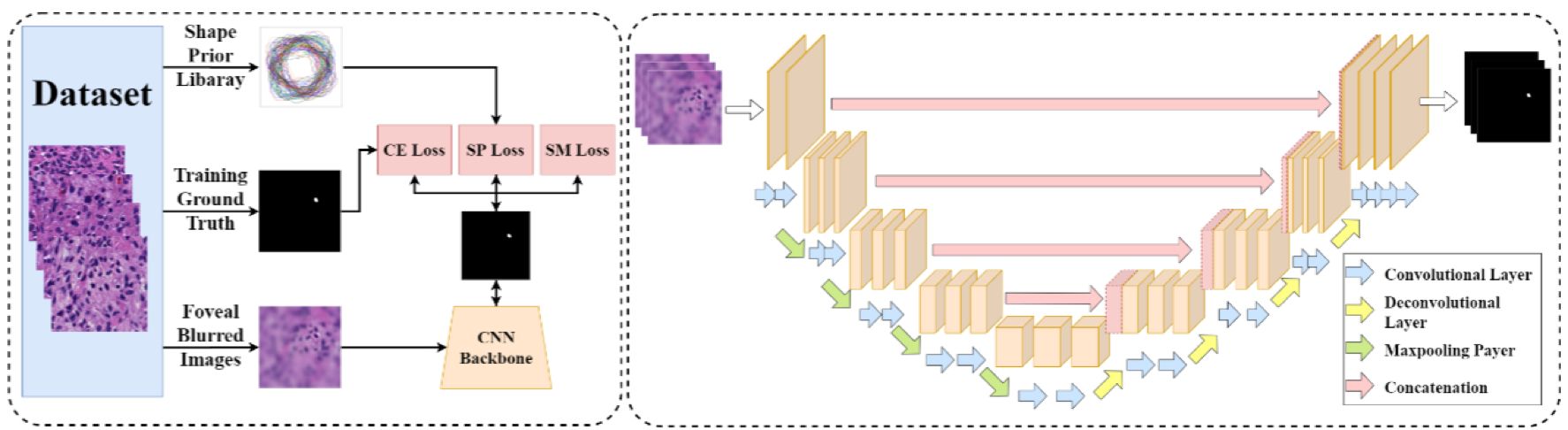
Pathology image analysis is a crucial step to support accurate cancer mechanism research, disease grading, diagnosis, and treatment planning. Although there is a high demand for precise nuclei morphology analyses, nuclei segmentation, the prerequisite step, mostly is completed manually by pathologists in practice. While the newly emerging deep learning algorithms can help automate the process, the lack of sufficient pathology training images with well-annotated ground truth in most tissue-based biomedical research inevitably limits the performance of deep learning systems. In this study, we propose to address this problem by a convolutional neural network with foveal blur, a blurring algorithm mimicking human vision, that enriches datasets with multiple local nuclei regions of interest derived from large pathology images for enhanced nuclei segmentation performance. As the convolutional neural network is not able to explicitly learn the nuclei shape, we propose a human knowledge boosted deep learning system by inclusion to the convolutional neural network new loss function terms capturing shape prior knowledge and imposing smoothness constraints. The high segmentation accuracy and competitive processing speed of our proposed method from three independent datasets suggest its promising potential for automating histopathology nuclei segmentation in biomedical research and clinical settings where efficient high-quality nuclei segmentation are demanded.
Source code:
https://github.com/jkonglab/Foveat_Cell_Segmentation
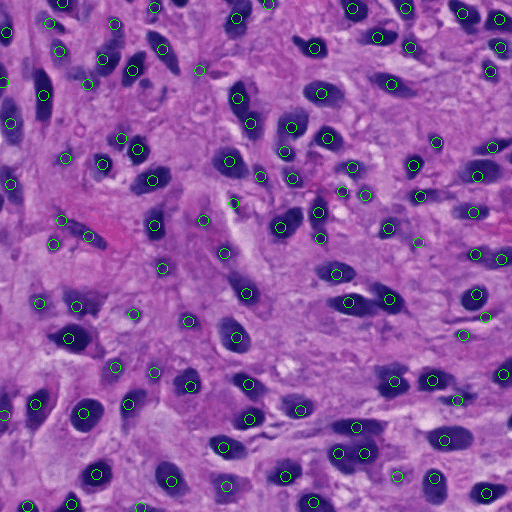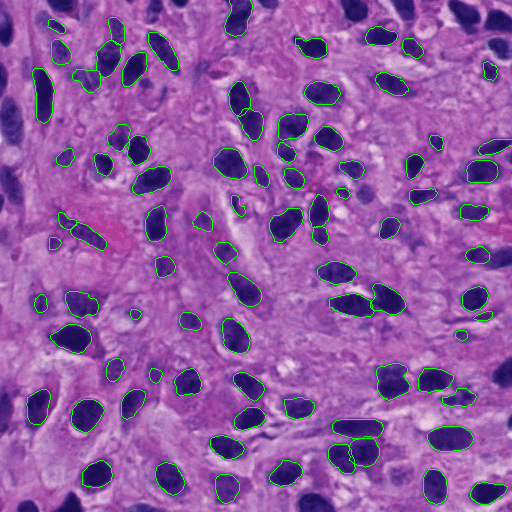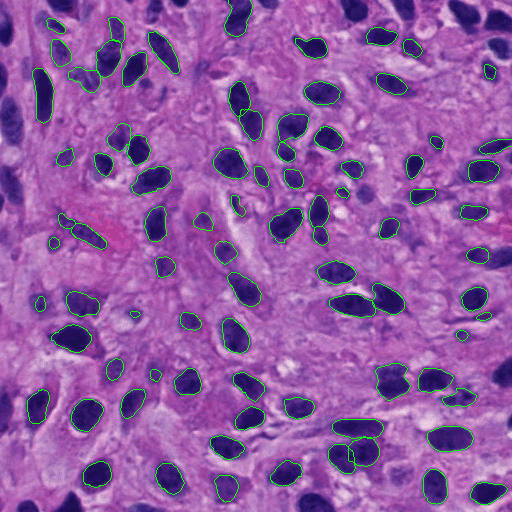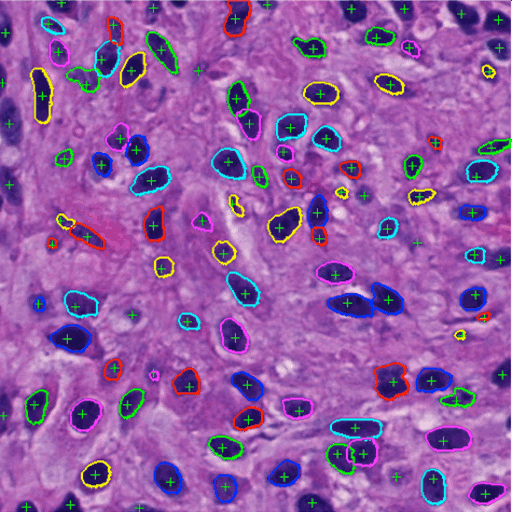
We have developed a complete and fully automated method for simultaneous segmentation of multiple nuclei in histology images with mutual occlusion. This method framework consists of a seed detection algorithm for nuclei contour initialization, and an integrated contour deformable model that incorporates region, shape, and boundary information. The developed nucleus seed detection algorithm utilizes joint information of spatial connectivity, distance constraint, image edge map, and a shape-based voting map derived from eigenvalue analysis of Hessian matrix across multiple scales. In the resulting nuclei segmentation step, a sparse representation-based shape term is introduced to better guide nuclei contour convergence with shape prior information from the shape library. All shape representations are clustered with an L1 graph-based manifold learning method to establish a concise and representative shape prior library. A dynamic nuclei occlusion term is proposed in the level set-based variational model to dynamically deal with occlusion events involving a variable number of nuclei. In-detail theoretical derivation for level set functional optimization and offer a solution to optimize the loss function. A numerical optimization algorithm is used to iteratively search for the desired level set functions. Experiments on histopathologic images produce better results as compared with other state of the arts, suggesting the effectiveness of our detection and segmentation approach for cell analysis with histopathologic images
Demo:
https://math.gsu.edu/jkong/research_cell_segmentation.html
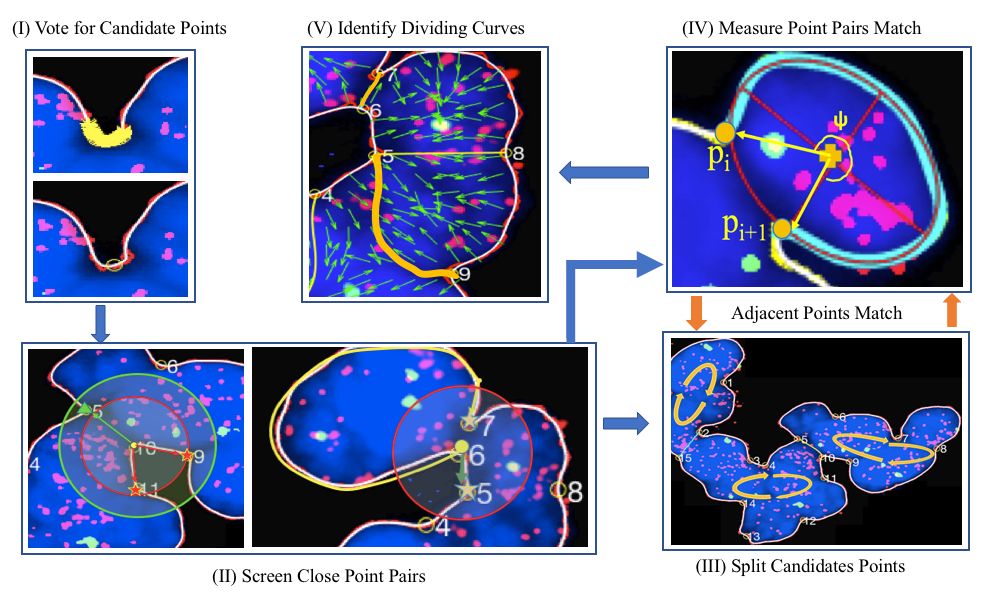
Highly clumped nuclei captured in fluorescence microscopy images are commonly observed in a wide spectrum of tissue-related biomedical investigations. To ensure the quality of downstream biomedical analyses, it is essential to accurately segment clustered nuclei. However, this presents a technical challenge as fluorescence intensity alone is often insufficient for recovering the true nuclei boundaries. We propose a segmentation algorithm that identifies point pair connection candidates and evaluates adjacent point connections with a formulated ellipse fitting quality indicator. After connection relationships are determined, we recover the resulting dividing paths by following points with specific eigenvalues from the image Hessian in a constrained searching space. Both qualitative and quantitative experimental results suggest that our algorithm is promising for dividing overlapped nuclei in fluorescence microscopy images widely used in various biomedical research
Source code:
https://github.com/jkonglab/Fluorescent_Nuclei_Segregation
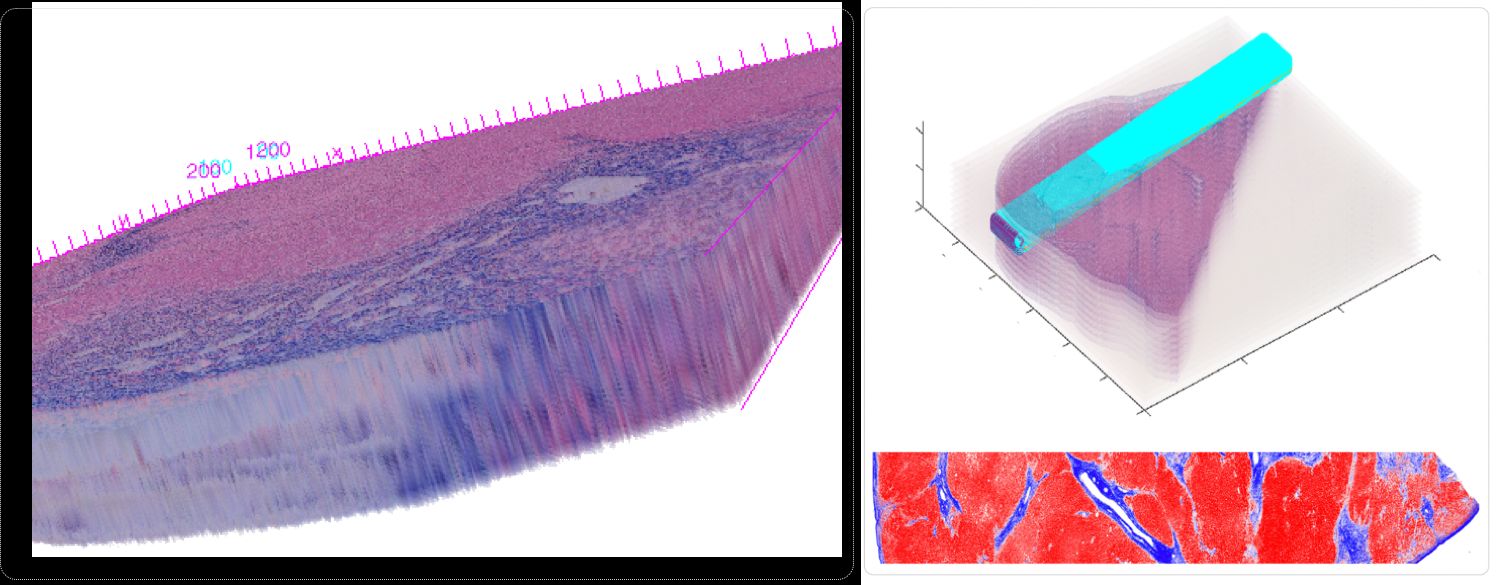
In spite of serving as the "gold-standard" for diagnosis of hepatic fibrosis and cirrhosis, the liver needle biopsy is known to include inaccurate diagnostic information, primarily due to the inexorable sampling bias. However, there is lack of method to gauge such sampling bias as it is infeasible to take a large number of needle biopsies from the same patient. To develop a computational analysis pipeline to enable needle biopsy sampling bias investigations, we construct a 3D virtual liver tissue volume by spatially registered high resolution Whole Slide Images (WSIs) of serial liver tissue sections with a novel dynamic registration method. With spatially aligned serial liver WSIs, we further develop a Virtual Needle Biopsy Sampling function (VNBS) that mimics the needle biopsy sampling process with a physical needle in current clinical practice. Such a virtual sampling function is applied to the reconstructed digital liver image volume for multiple times, each at a distinct tissue location and angle. Additionally, we develop an analysis pipeline to quantitate Collagen Proportionate Area (CPA) in all resulting needle biopsies. All sampled biopsies are manually graded by the Scheuer and Ishak staging methods. With such domain annotations and machine-based collagen fiber quantification results, we investigate the inter- and intra-biopsy variability by the pathology stage score and CPA with a set of 3D virtual needle biopsies and their internal 2D cross sections. Our experimental results suggest the developed computational pipeline can recover the substantial presence of needle biopsy sampling bias in both pathology stage scores and liver CPAs.
Conclusions: The developed virtual liver needle biopsy sampling method provides a new avenue for investigating needle biopsy sampling bias with 3D virtual tissue volumes. Although the developed working pipeline specifically investigates the sampling bias in liver needle biopsy in this study, the presented analysis methods can be tailored to a large scope of other tissue based disease diagnoses where the needle biopsy sampling bias substantially affects the diagnostic results.
Source code:
https://github.com/jkonglab/3D_Liver_Biopsy_Sampling
Demo:
https://dplab.gsu.edu/demo